
Biologists have long deemed the task of predicting evolution a lost cause due to the complexity of biological systems and their interactions with the environment. An increasing number of observations of repeated evolution have however recently challenged this view suggesting that it may be possible to make short-term evolutionary forecasts.
Why Predict Evolution?
The predictability of evolutionary processes is of fundamental importance to biology, but it could also have applications in major areas related to human health, including evolution of new pathogens, antimicrobial resistance, vaccine efficacy and personalized cancer prognosis and treatment. Other applications could be possible in predicting organisms’ ability to adapt to climate change, gene drives, design of microbial communities and synthetic biology.
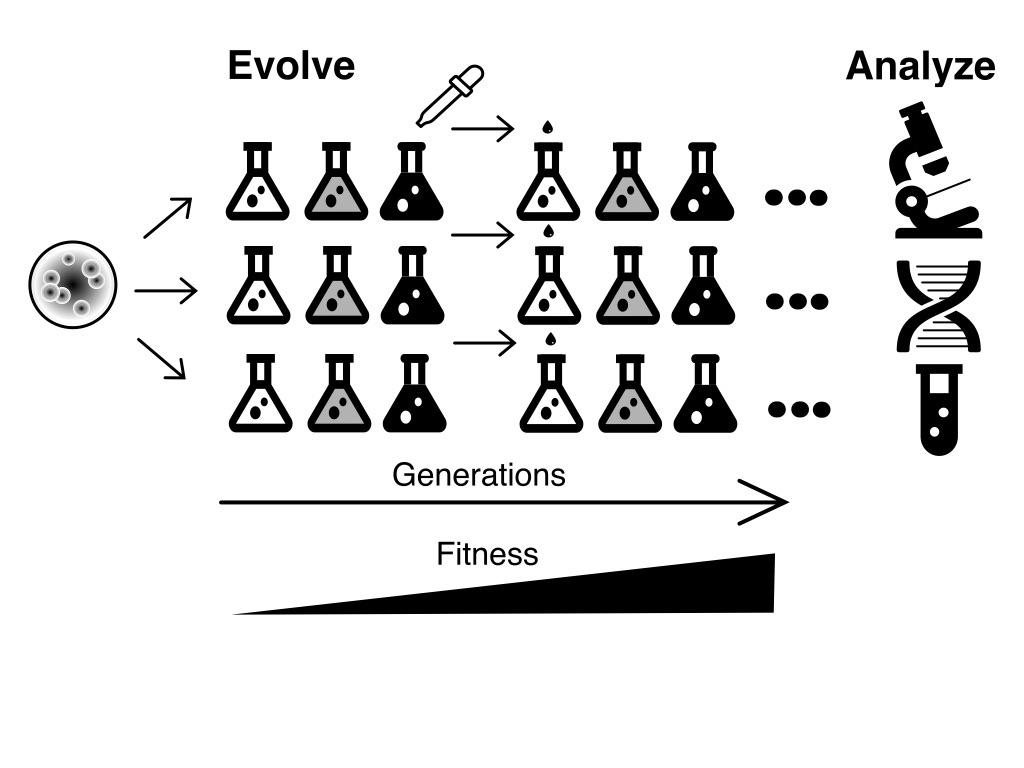
Experimental Evolution
By allowing bacteria to adapt to a new environment in the laboratory we can study evolution in real time and beneficial mutations can be identified by DNA sequencing. Bacteria can be maintained at large population sizes and have short generation times so evolution can be studied over many generations. A fossil-record can be maintained in a non-evolving state in the freezer allowing fitness comparisons between evolved and ancestral strains. Genetic tools are available for reconstructing and combining mutations to determine their phenotypic effects.
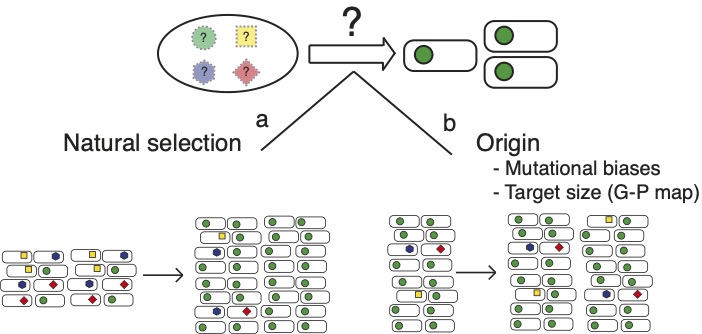
What Makes Evolution Repeatable?
The causes of repeated evolution is not trivial to determine even in laboratory populations. Possibly, only one gene can be mutated to cause a particular adaptive phenotype and then repeated mutations in this gene is the only possible solution. Alternatively, there are several genes that can be mutated, but one give rise to phenotypes with higher fitness and is favoured by natural selection. It is also possible that mutations in several genes give the same high fitness, but that mutations in some genes are more common. A higher mutation rate to a phenotype for a gene could be either because of a larger mutational target size or mutational biases between sites.

Model Systems for Predicting Experimental Evolution
We currently use two model systems where we predict experimental evolution using experimental data combined with computational predictions followed by experimental tests and updated forecasting models. The first focus on biofilm formation at the air-liquid interface during static growth for different Pseudomonas species. For our second model system we predict the evolution of multidrug resistance in Pseudomonas aeruginosa. Mutations causing increased biofilm formation and multidrug resistance commonly evolve in humans during infections and we continually evaluate the medical relevance of our results. Using a third model system, focusing on the evolution of macroscopic multicellularity in bacteria, we investigate the role of phenotypic plasticity in the evolution of new phenotypes.